Cellular and molecular biomechanics, microscopy
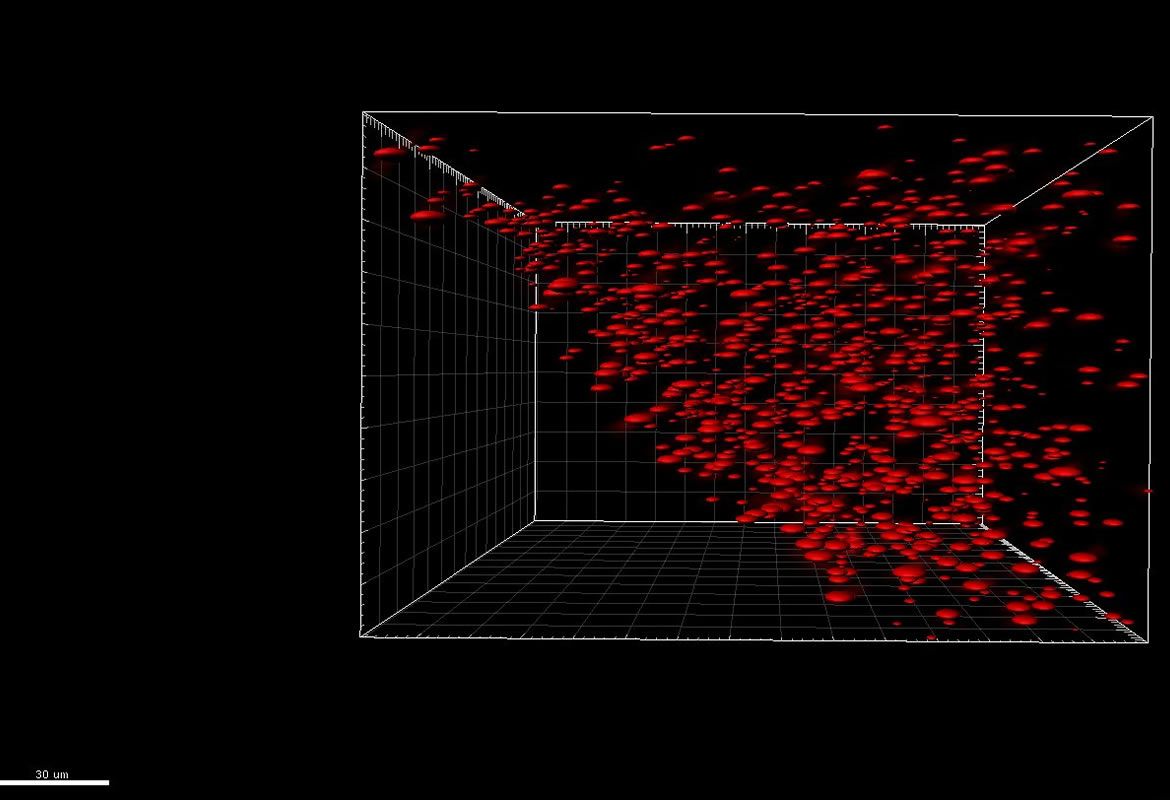
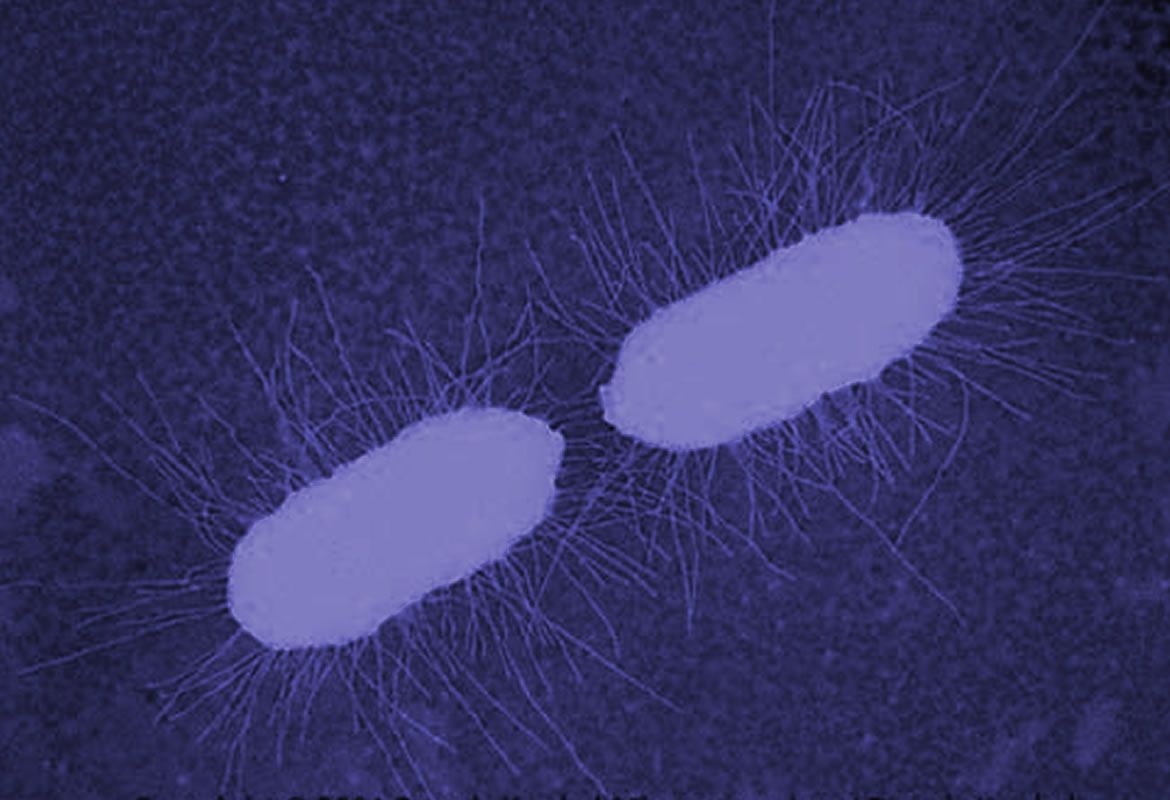
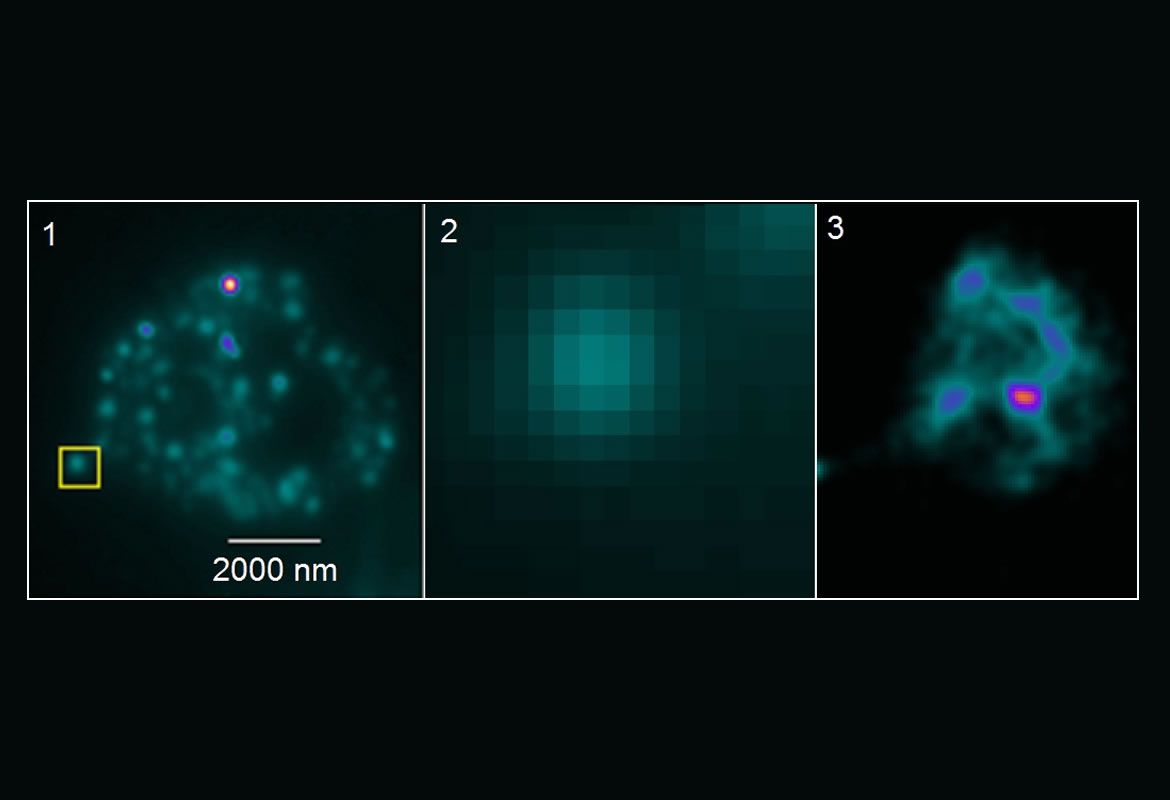
Bacteria have appendices similar to hair named pili, which allow the adhesion of bacteria cells to human cells in flow conditions. In E. coli, this adhesion promotes the generation of infections such as cystitis. In previous research, it has been shown that pili can extend like elastic strings to limit the effect of big forces caused by the fluid flow in the adhesive end of pili. In our previous projects we have shown that the molecule FimH, in charge of the adhesion between pili and the surface, presents amazing adhesive properties. Besides, we have evidenced that the pili has a remarkable property: it can have a greater adhesion under tension than without tension. This counter-intuitive property is called catch bond, quite similar to the finger traps found in kid’s toys.
In a collaboration project with the Mechanical Engineering department, we want to run simulations and experiments on multiple levels to understand the way in which the fluid, pili and FimH properties affect bacterial adhesion.