Membrane Biophysics
We have developed a strategy of multiple experimental techniques, simulations and theoretical analysis to study the thermodynamics, mechanical and structural aspects of biomembranes. Our purpose is to explore how the physico-chemical properties of biomembranes play a role in the regulation of the physiological processes of the cell. We use fluorescence spectroscopy and FTIR as experimental tools to elucidate physical aspects of membranes such as packing level, lateral organization and permeability. We complement this with diverse microscopy techniques (fluorescence microscopy and atomic force microscopy) to study structural and mechanical properties of membranes at a microscopic and nanoscopic level. This work is carried in membrane model systems such as liposomes, supported membranes and giant unilamelar vesicles (GUVs). We translate this knowledge to a cellular level with biological models in bacteria and eukaryotic cells.
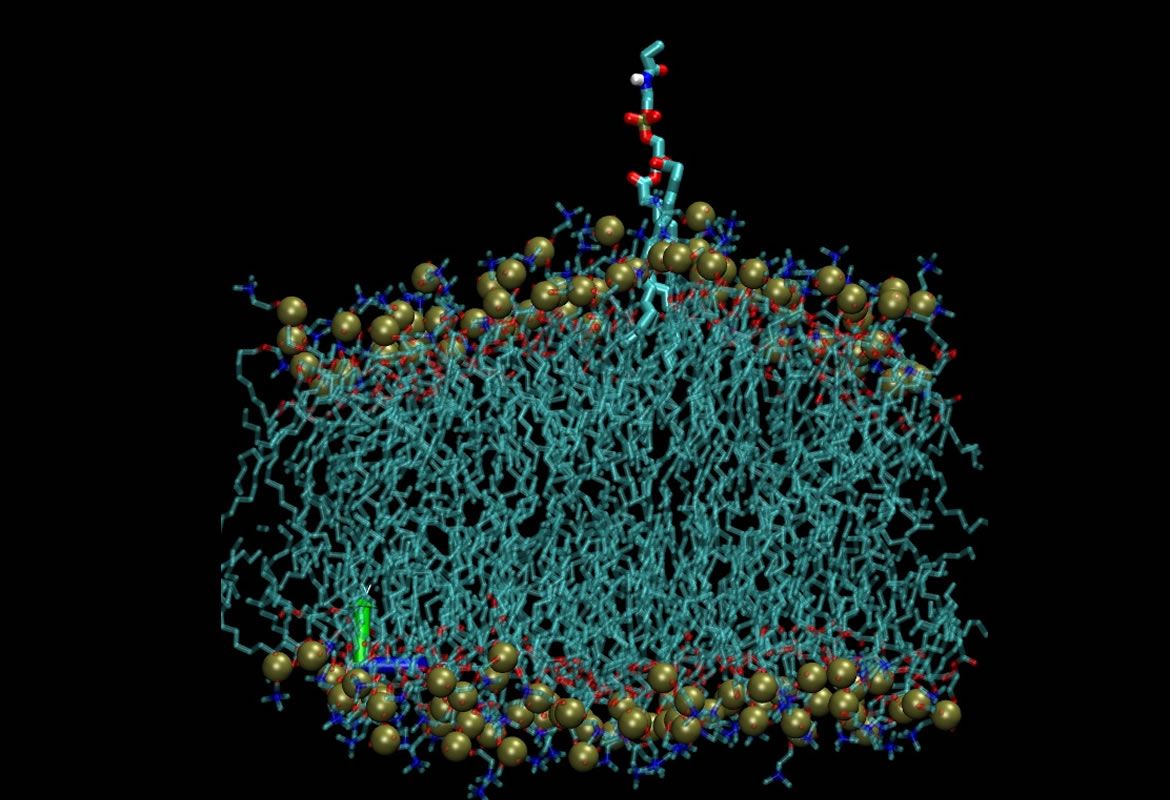
Molecular dynamics of membranes to study their mechanical properties
We have developed a molecular dynamics technique to determine the elastic properties of different lipid systems. We use umbrella sampling to determine the Potential Mid Force (PMF) and to study deformation profiles in order to understand how the presence of different membrane modifiers (lisolipids, detergents, peptides) affect these mechanical properties.
Rafael Clavijo, Juan David Orjuela, Colaboradores: Gilles Pieffet, Günther Peters, Roland Faller
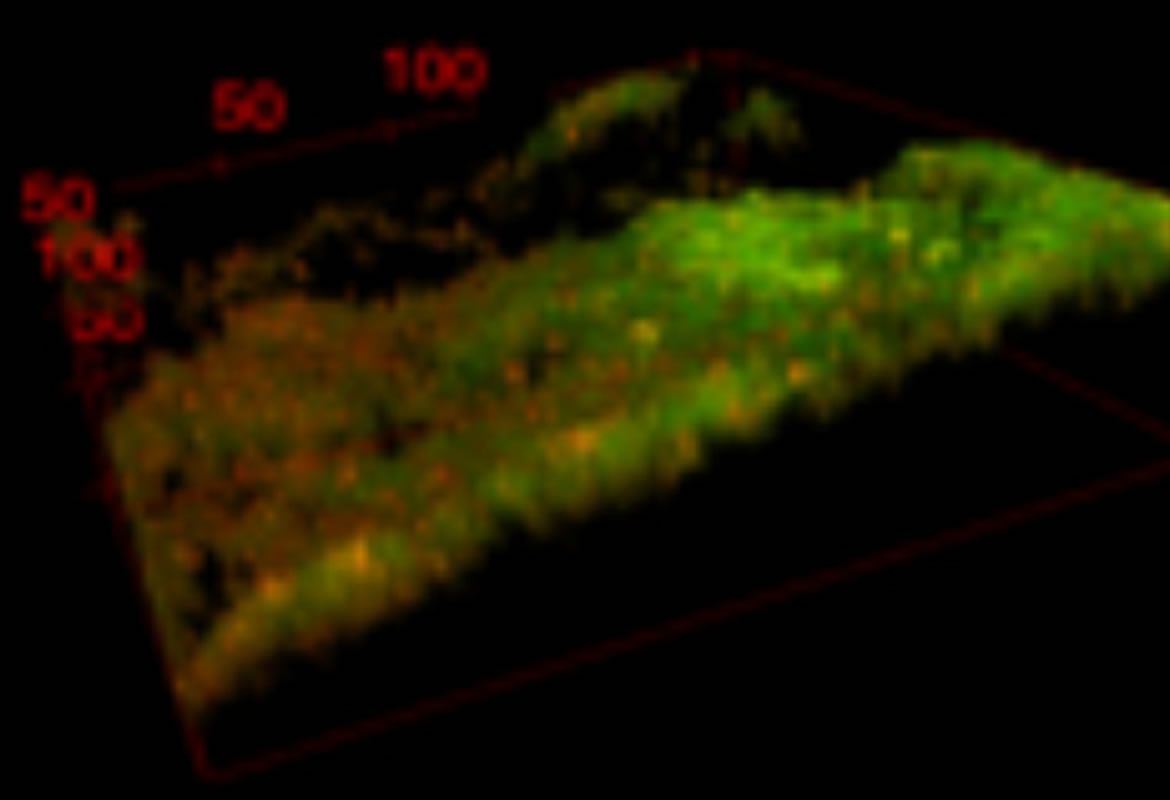
Relation between membrane lipid composition and antimicrobial peptide susceptibility in S. aureus
Membranes affects membrane fluidity and modifies antimicrobial peptide susceptibility. Bacteria have the capacity of modifying lipid composition to adapt to new growth conditions. We are interested in knowing the membrane lipid composition in biofilms and the susceptibility to different antimicrobial peptides.
María Isabel Pérez López, Steve Trier

Study of survival strategies of S. aureus biofilms exposed to membrane-active antimicrobial peptides
The action mechanism of antimicrobial peptides results in the destabilization of the biofilm structure. Biofilm sections are affected in different ways by the action of antimicrobial peptides, depending on the exposure of cells to the peptide. Usingconfocal microscopy, we want to determine the biofilm section that presents the greatest damage due to peptide activity, as well as defining sub-populations within the biofilm that show the greatest susceptibility to the antimicrobial peptide.
María Isabel Perez, Steve Trier

Membrane fluctuation analysis in giant unilamelar vesicles
Membranes are essential constituents of any living organism. Their mechanical properties determine diverse aspects of physiological importance for the cell, such as stability and external factor resistance. Using Vesicle Fluctuation Analysis (VFA), one can infer the bending elastic constant and the rigidity of lipid membranes. We are interested in studying the changes in these constants under different conditions, such as the presence of charges or cholesterol.
Rudy Mendez in collaboration with Andres Gonzalez Mancera
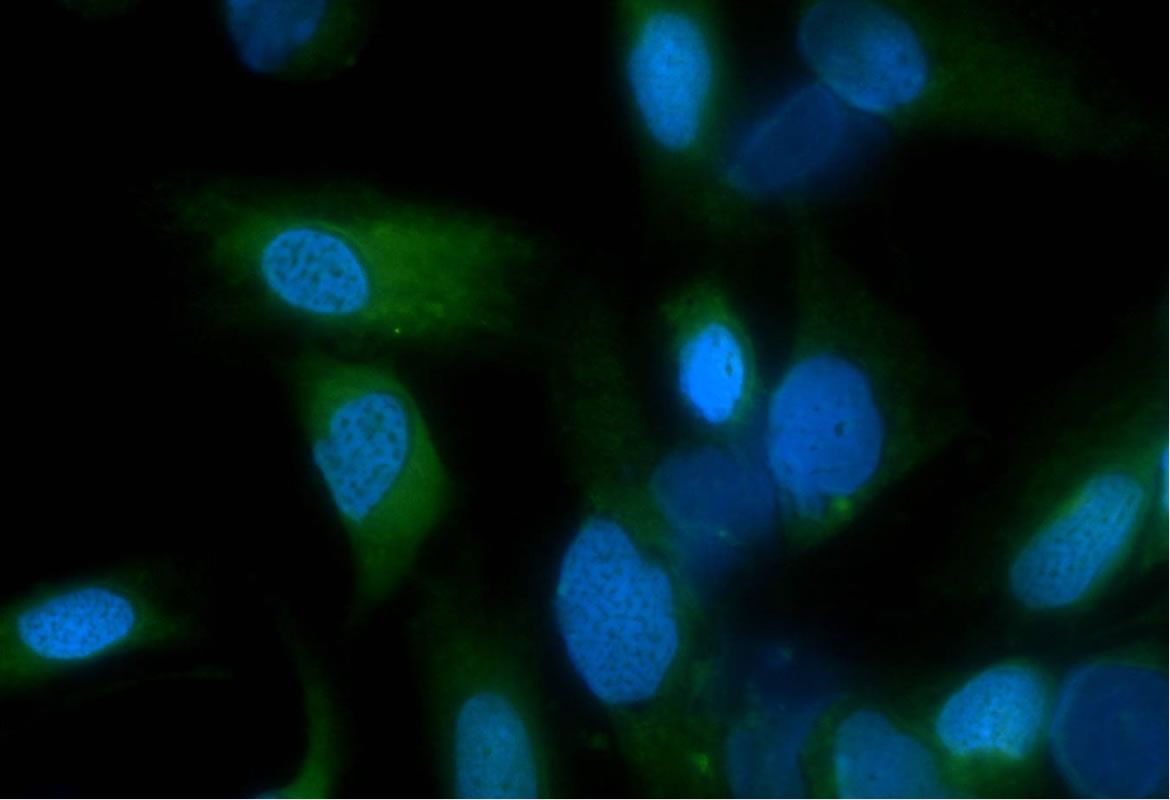
Liposome trapping processes by tumor origin cell lines
In this project we want to identify specific receptors to increase the trapping percentage of liposomes. Additionally, we are interested in analyzing how the liposome-membrane composition regulates catch and release processes of the liposomal content in tumor origin cell lines.